Bioimaging & Embryology
- Deputy Head:
- Research associate:
- Laboratory technicians:
The most prominent focus of the unit is to provide functional morphological analysis of phenotypes in adult mice and rat models and during their embryonic development. The microCT technology provides the best cost effective approach for 3D visualization of phenotypes and the unit provides full data analysis platform 3D data processing. Besides 3D imaging, the unit is equipped with a whole body imaging system that is suitable for imaging of fluorescence and bioluminescence reporters in mice and rats in vivo and is very advantageous especially for imaging of cancer models derived from cancer cell lines or PDX. For non-invasive imaging and cell labelling the luminiscent and fluorescent lentiviral reporters are available. Apart from cancer cells, the physiological processes like inflammation, kidney function or specific enzyme activity can be also non-invasively imaged. Beside stated imaging modalities, the unit also provides experience in functional assays on primary cells or their isolation for multiOMICs technologies. The embryological tissues can be dissected and primary cell lines or organ cultures can be established as well as immortalized cell lines from knock out phenotypes can be delivered. These approaches accelerate the research of mutants with embryonic lethal phenotypes.
Comprehensive morphological and functional characterization of animal models by whole-body imaging systems in vivo and ex vivo.
Standard Services MicroCT
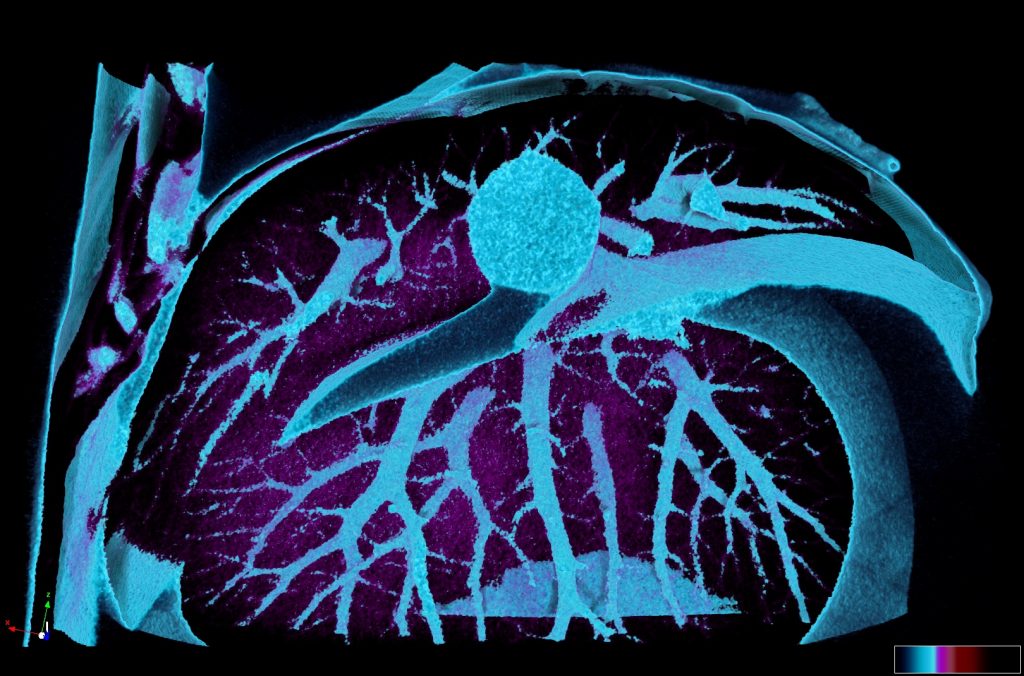
Our microCT in vivo scanner provides sensitive and high resolution 3D imaging based on X-ray projections with voxel size 9 – 35um. Our imaging set up is suitable either for in vivo or ex vivo imaging. In vivo scans can provide comprehensive 3D visualization of bones and other mineralized tissues like teeth, but also quantification of total body fat mass and lean mass (alternative to DEXA analysis). MicroCT also provides fast imaging mode for in vivo visualization of cardiovascular system and kidneys after application of contrast agents.
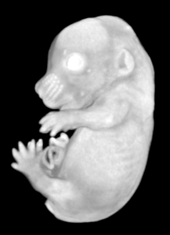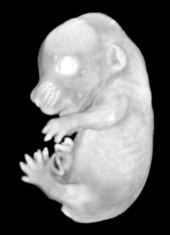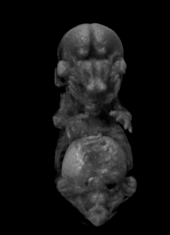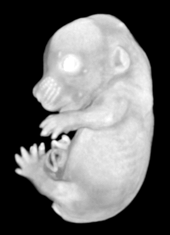
Ex vivo scanning mode can be used for higher resolution imaging or use of contrast agents for imaging soft tissues (liver, kidney, hearth, neuronal tissue) or imaging fixed embryos (E7.5 – E18.5) with resolution down to 0.5um with use of appropriate contrast agents (Iodine, PTA). For complete microCT analysis software post-processing of projection data is necessary, especially 3D reconstruction and further segmentation for visualization of morphological phenotypes. Data can be presented as static images and/or as animated 3D reconstruction movies.
Software tool box: CTvox, CTanalyser, Imaris, Amira, ITK-snap, 3D-slicer
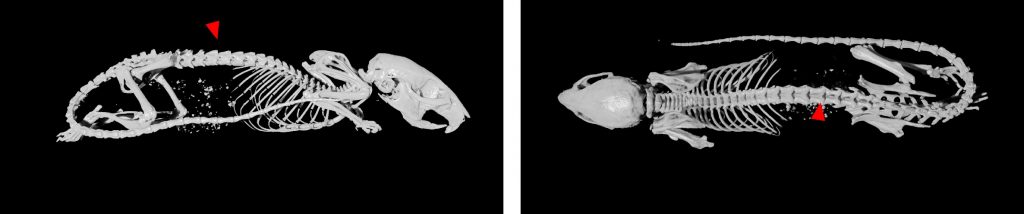
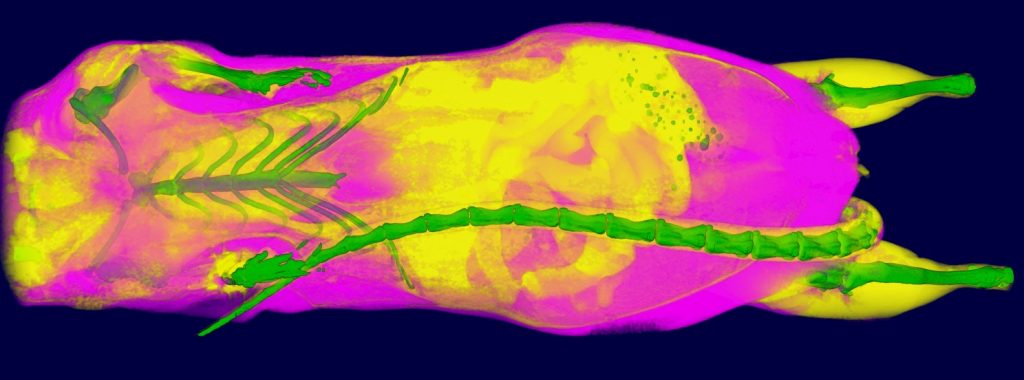
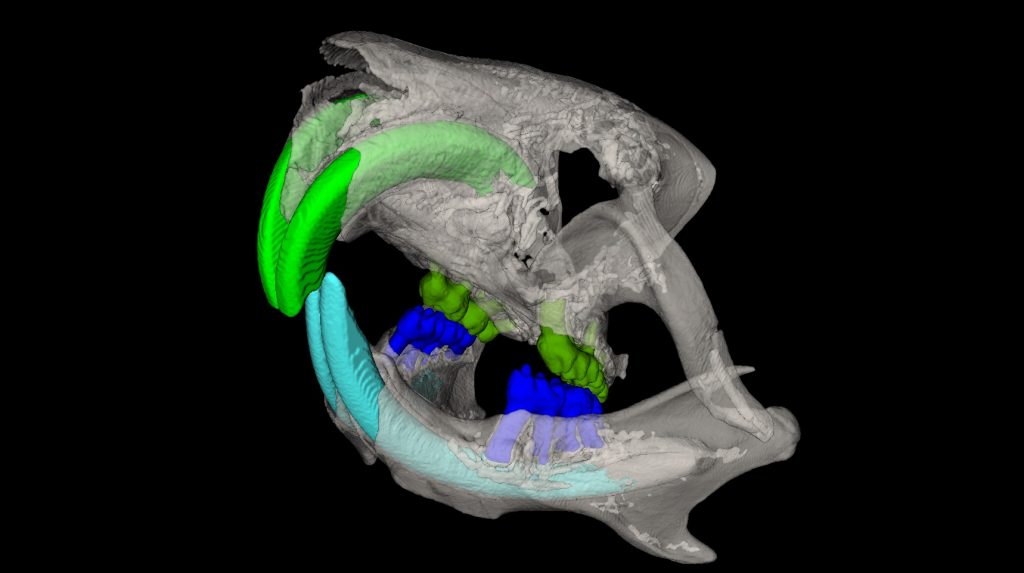
Standard Services Radiography (x-ray)
2D radiography is the best for fast analysis of mineralized/hard tissues, with combination of rotation stage can be also combined in 3D projection. This imaging modularity is mostly recommended as background image for other modularities (luminescence/fluorescence). Combination of optical imaging and X-ray is very suitable for anatomical annotation of fluorescence or luminescence signals.
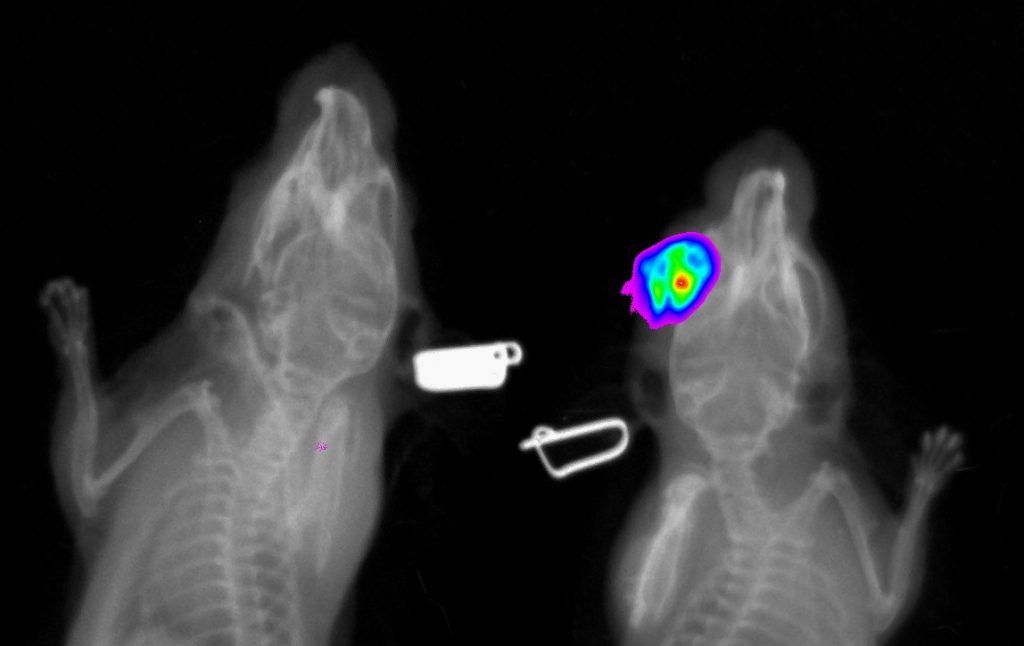
Standard services Whole Body Fluorescence Imaging
Our device is very suitable for imaging of reflected fluorescence signals which can be used for in vivo or ex vivo imaging. Even though the ex-vivo imaging provides better signal resolution, we focus mostly on application of in vivo approach in most cases. In vivo fluorescence imaging provides unique opportunities for visualization and quantification of pathological processes like inflammation, kidney function or tumour progression in longitudinal analysis.
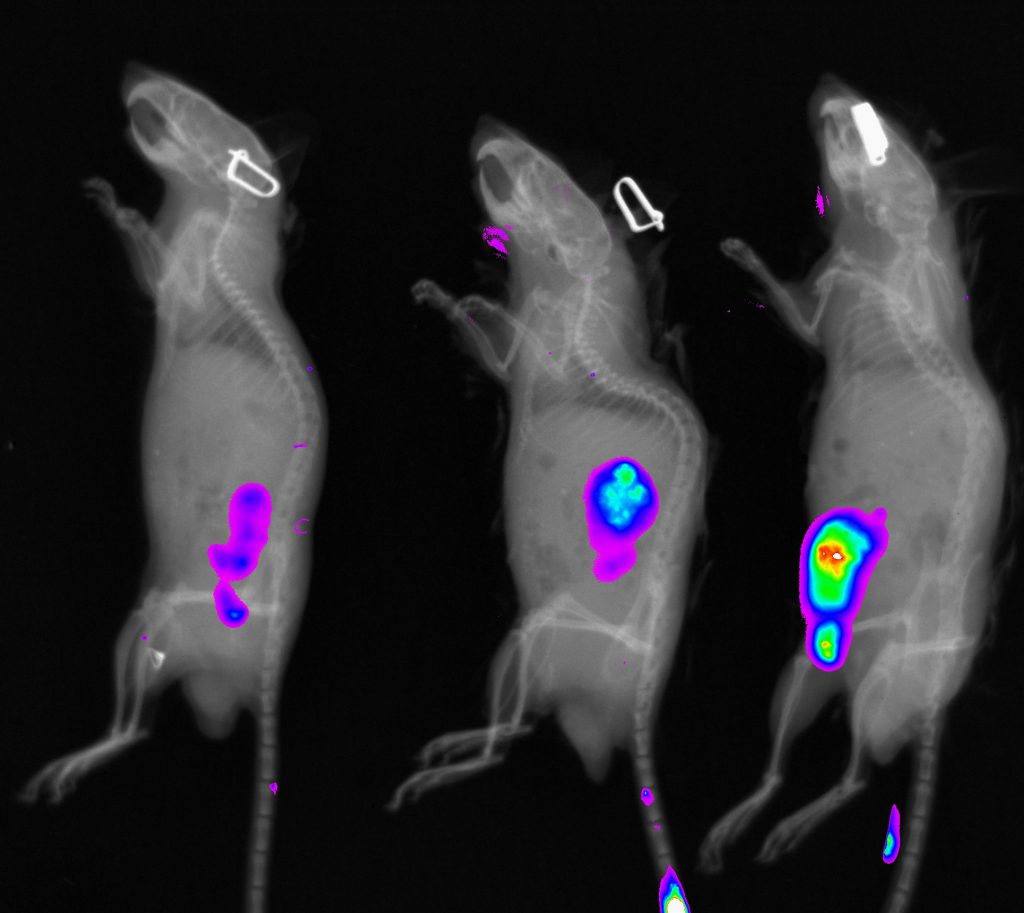
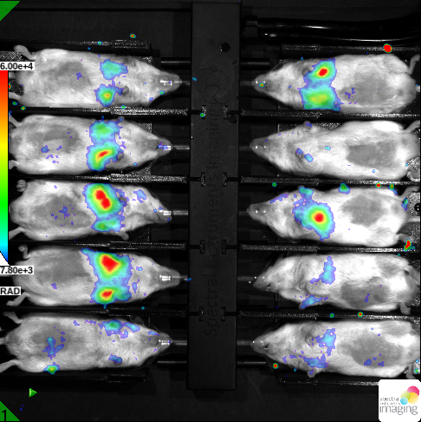
Standard Services In vivo Bioluminiscence Detection
Our device includes highly sensitive camera for detection of emitted photons based on enzymatic activity like luciferase or peroxidase. This approach is suitable for in vivo imaging with emphasis on high sensitivity. This imaging is frequently used for inflammation, tumour progression, metastasis or cell homing experiments. We have successfully published our imaging protocol in the study of genetic regulation of DSS colitis. We offer our knowledge for screening of potential role of given genes in the gut regeneration processes by non-invasive imaging setup for longitudinal monitoring of healing process.
Standard Services Tissue Clearing and Large Z-stack Optical Sectioning
We can offer our experience with tissue clearing and whole-mount imaging protocols employed predominantly for cre dependent fluorescence reporters (R26mT/mG) since a standard data can be presented as volume rendered static images or animated 3D reconstructions.
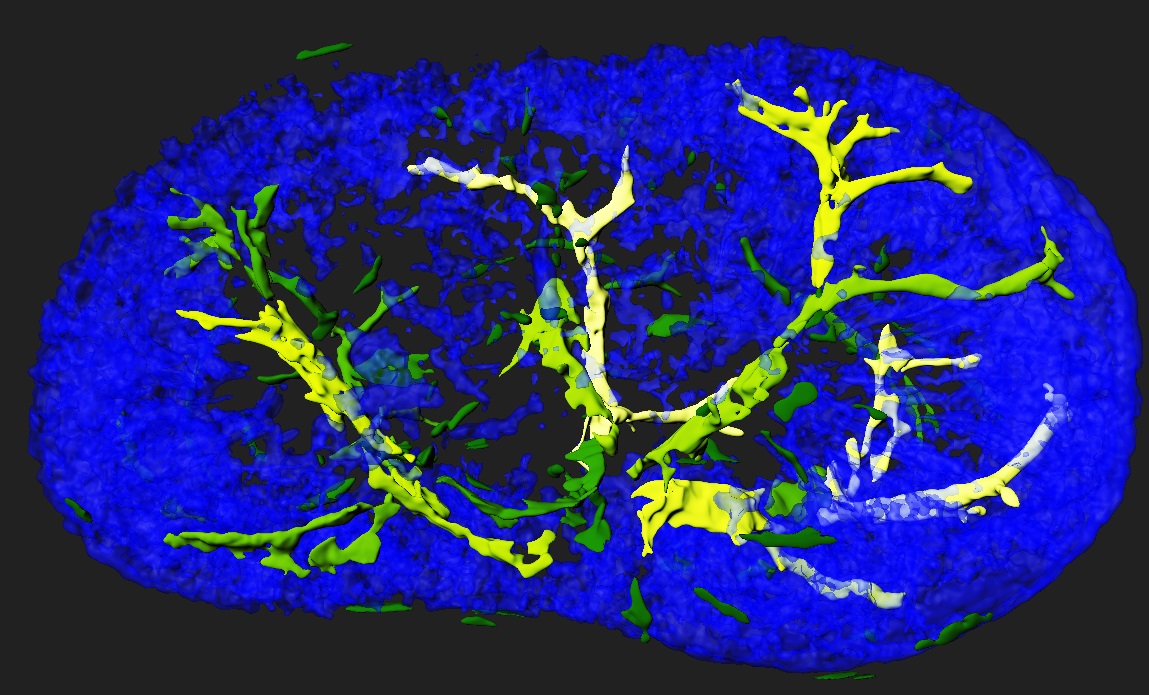
Standard services Histological Tomography
In collaboration with histology unit we can offer unique 3D visualization from serial histological sections. In this procedure, the entire organ or embryo is sectioned on histological slides, processed for hematoxylin or any other histological staining, every single section is digitalized and then 3D reconstructed in Voloom software. Datasets can be also processed in Bitplane Imaris for state of the art quantification analysis.
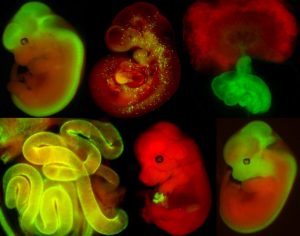
Standard Services Embryology Services
Whole Embryo and Placenta Isolation/Dissection
Embryos can be dissected from E7.5 in following half day intervals up to E18.5. The genotyping of embryos and resorptions are among standard procedures and harvested material can delivered up to request (fresh, frozen or fixed). The embryos can be obtained also from conditionally inactivated gene models, where gene ablation design is discussed first. Suitable combination of genetic driver and reporter (see examples of our availabe Cre drivers crossed to R26mT/mG below) can be also perfomed in order to visualize and follow specific cell populations.
From 12.5 dpc onwards, the mouse skeleton can be visualized using the histochemical stains alcian blue (which stains the cartilaginous skeleton) and alizarin red (which stains mineralized tissues).
Staining for ß-galactosidase Activity
The most of knockout mouse lines generated via KOMP/ EUCOMM embryonic stem cell resources have introduced the lacZ reporter gene driven by the endogenous regulatory elements of the targeted gene. These or other appropriete strains carrying a lacZ expression construct can be used for whole-mount ß-galactosidase staining or staining of sections. Both transgenic embryos (placentas) and adult organs can be stained. Regarding the prenatal period, from E13.5 the staining is performed on dissected organs or frozen sections in order to minimize problems with substrate penetration in larger embryos. The principle of this method is histochemical staining for LacZ enzyme activity which can be detected in organs, substructures, and cells in which the gene is normally expressed. LacZ expression analysis is thus crucial for understanding of the physiological functions of a gene as well as for its role in the development or progression of diseases.
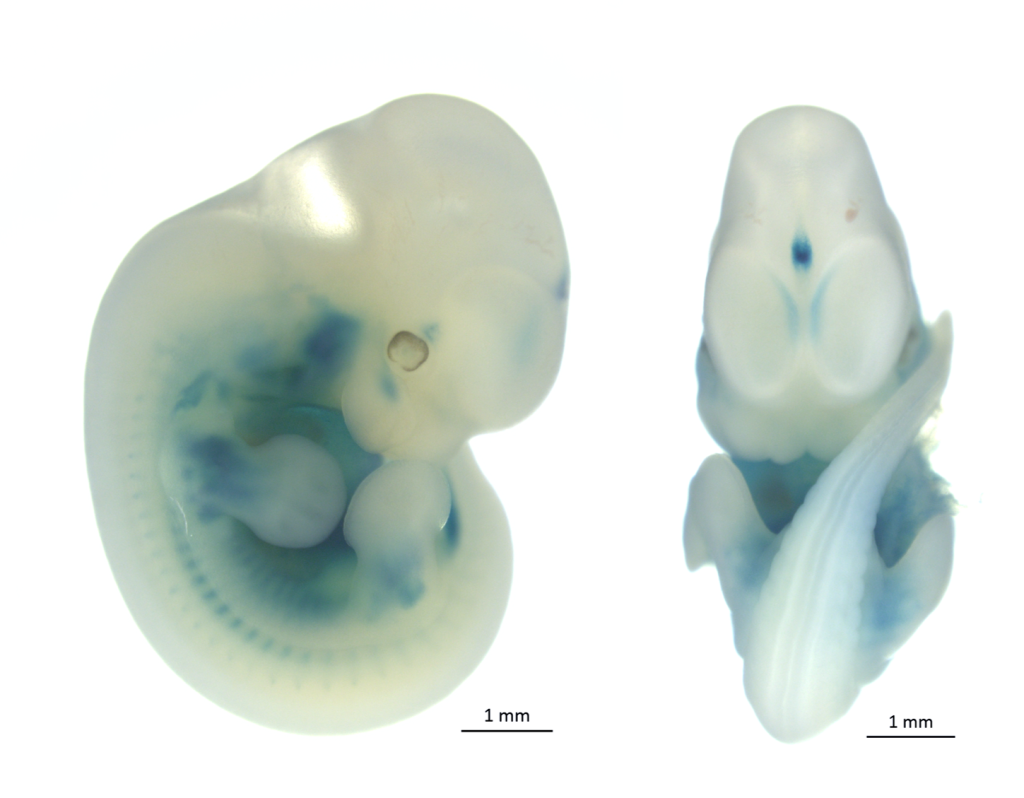
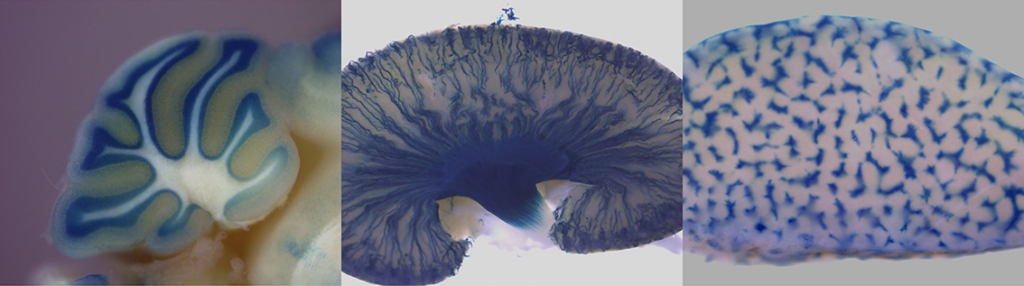
LacZ stained embryo and adult organs of KOMP/ EUCOMM generated strains.
Immunohistochemistry and In situ Hybridization Staining
Expression of RNAs or proteins can be detected using in-situ hybridization or immunohistochemistry respectively. Both can be performed either in whole mount or on sections and followed by 3D imaging methods.
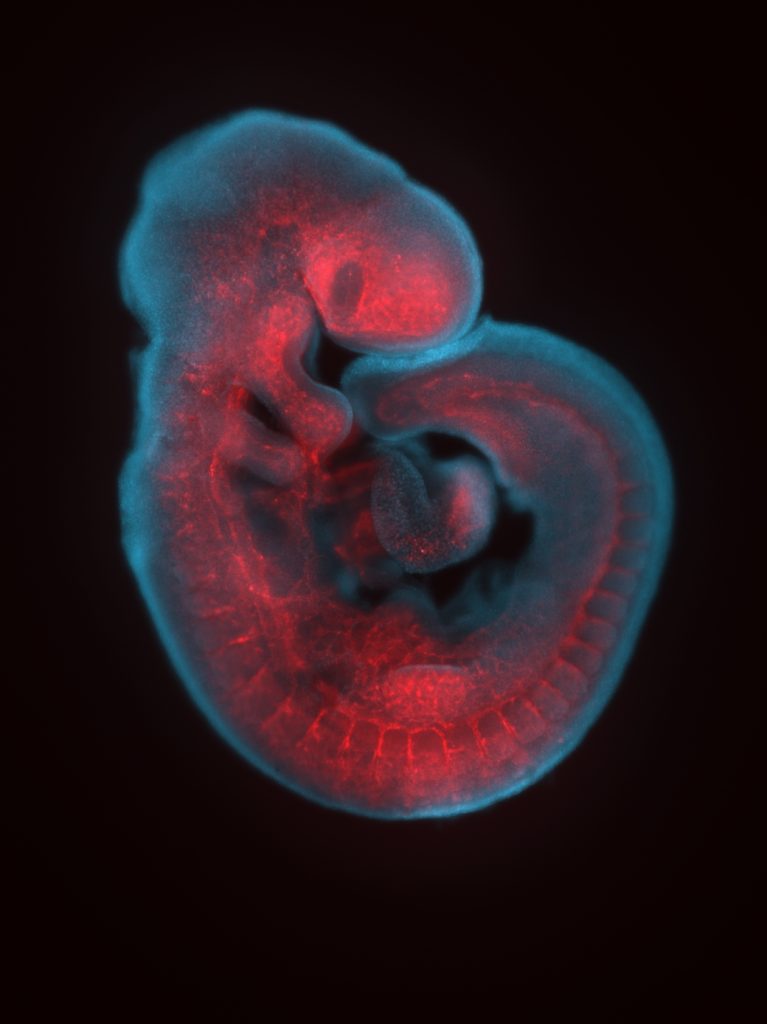
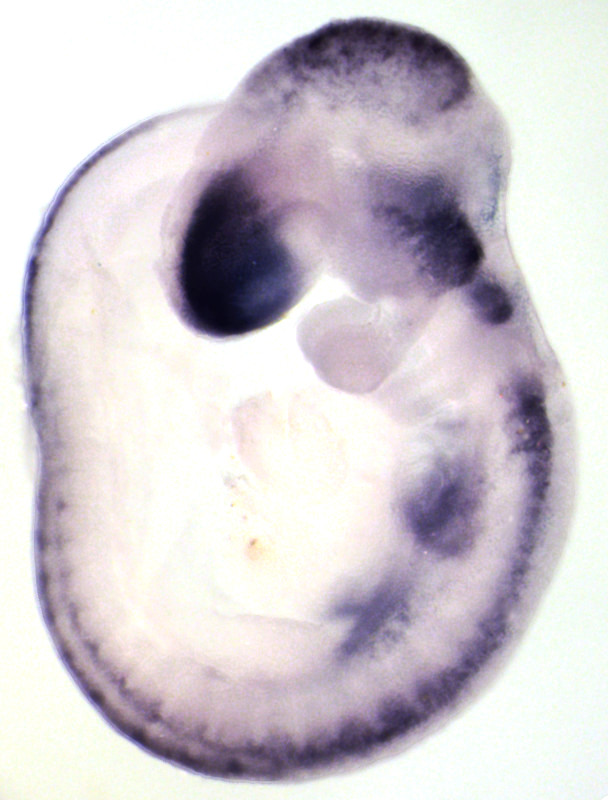
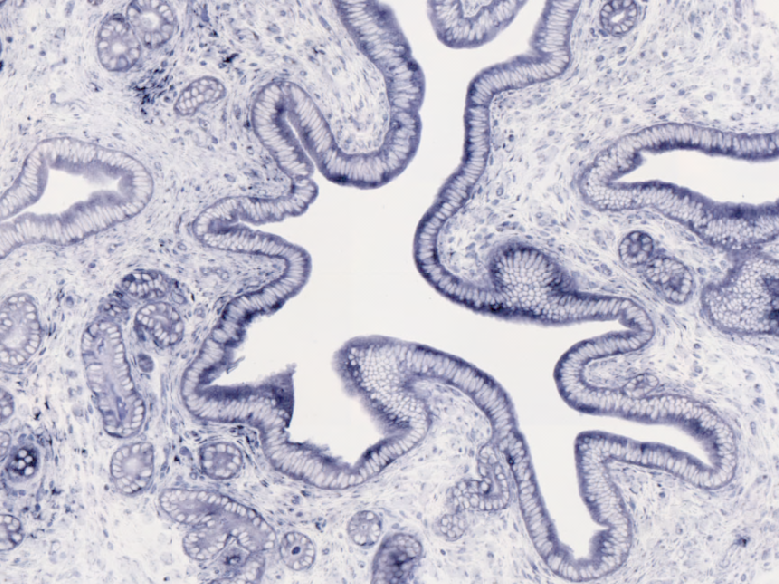
Whole-mount immunohistochemistry staining in E9.5 embryo and in situ hybridization staining in whole mount (embryo) and on section (adult ovary).
Histochemical Visualization of Embryonic Skeletons
From 12.5 dpc onwards, the mouse skeleton can be visualized using the histochemical stains alcian blue (which stains the cartilaginous skeleton) and alizarin red (which stains mineralized tissues).
Embryo Imaging and Image Analysis
We use multiple imaging modalities for detail phenotype analysis in embryos of all developmental stages. We use combination of standard and high resolution microCT techniques for detail morphology and anatomy analysis, further image segmentation approaches can be used for quantification and presentation of phenotype in direct publication quality. Another imaging modality is detection of fluorescence where we use multiple tissue clearing protocols (ScaleA2, Cubic, BABB, Clarity) in combination with deep tissue imaging techniques for large Z-stack optical sectioning (confocal imaging, Light Sheet microscopy, Two-photon confocal imaging and Structued illumination microscopy). The data can be presented as volume rendered static images or animated 3D reconstructions. For image analysis we can offer Imaris analytical station with appropriate modules for developmental biology (filament and particle tracking, Matlab extension).
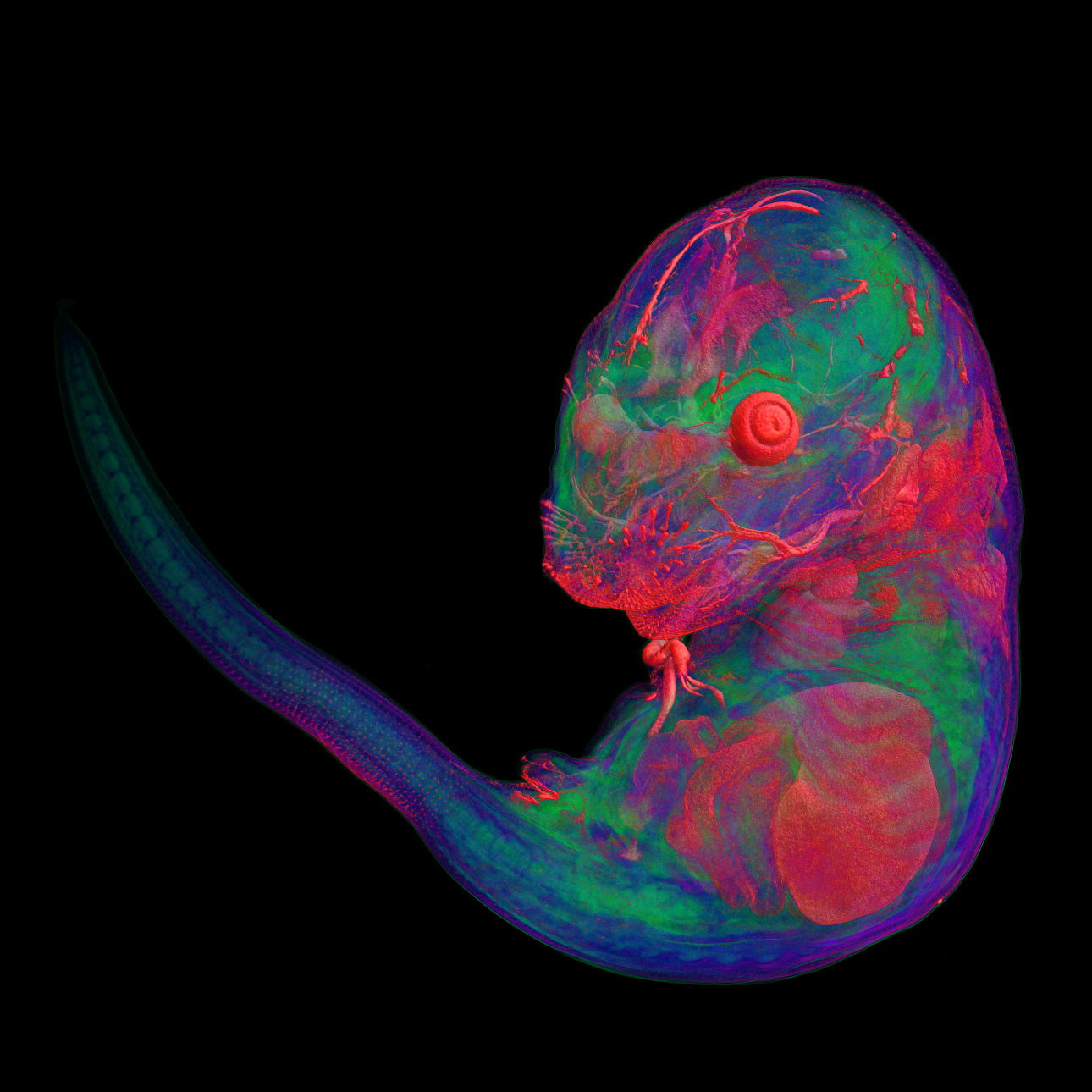
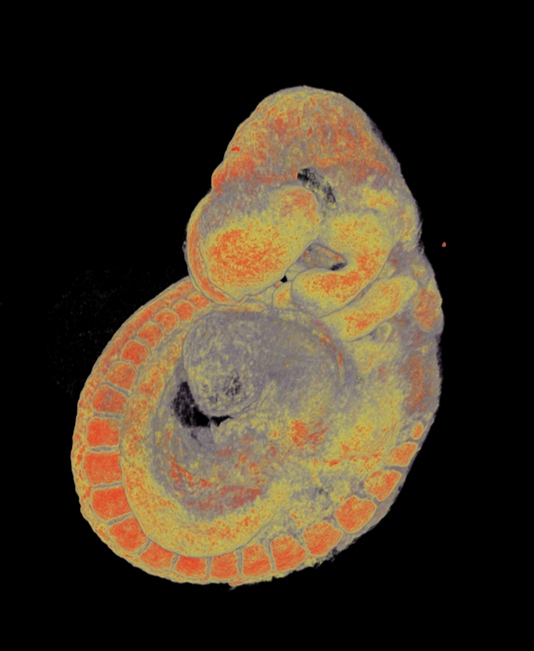
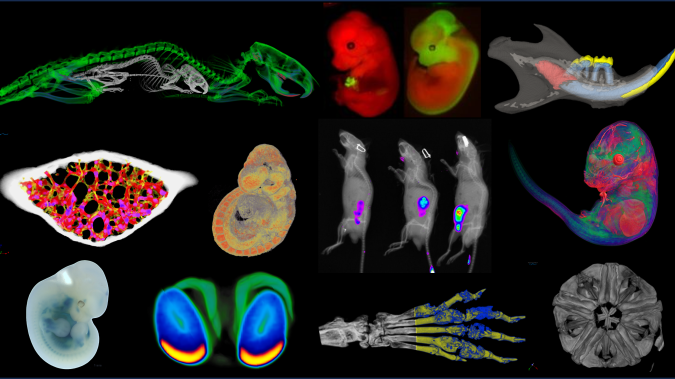
Pseudocolored microCT scans of mutant embryos harvested at E18.5 and E9.5.
Embryonic fibroblasts
Embryonic fibroblasts (MEFs) can be readily isolated from mouse embryos, standard stage of harvest is E13.5 but can be adjusted upon request
Technology platforms
Bioimaging unit was upgraded with the support from OP RDE projects CZ.02.1.01/0.0/0.0/16_013/0001789 – Upgrade of the Czech Centre for Phenogenomics: developing towards translation research and CZ.02.1.01/0.0/0.0/18_046/0015861 CCP Infrastructure Upgrade II in the years 2020 – 2022 and currently it is being upgraded from the OP JAC project CZ.02.01.01/00/23_015/0008189 Upgrade of the large research infrastructure CCP III.